LigerCat2
LigerCat2 is a search engine that presents an alternate interface for finding and accessing all prematurity-related articles and genes on PubMed for researchers and mothers of premature infants. Researchers can use LigerCat2 to cross-reference prematurity-related genes and articles. Mothers of premature infants can use LigerCat2 to view machine-generated summaries and relevant genes for key research articles in the prematurity literature.
Mentor: Dr. Neil Sarkar
Timeframe: 4 months
Scope of the project
During the summer of 2018, I worked with Dr. Sarkar at Brown's Center for Biomedical Informatics on LigerCat2, an extension of the original LigerCat Dr. Sarkar developed a few years ago. Dr. Sarkar initially introduced the idea of this project the previous summer as a way to streamline the exploratory phase of research. By creating a streamlined interface for aggregating genes from relevant articles and vice versa, researchers can avoid the distracting context-switching that happens when copying and pasting query results across multiple NCBI web pages.
Coming from a less-specialized background, I was also interested in how much people who were affected by prematurity in some way knew about all the research that was happening in this area. Where would a mother of a premature newborn turn to learn more about how prematurity would affect her child? In an age of self-service medicine, for better or for worse, anyone can look up information about nearly any disease. But how often do patients actually dive into primary literature? While working on LigerCat2, I wanted my work to at least begin to explore these questions.
To help manage the scope and clarify the deliverables of this project, I worked with Dr. Sarkar to develop a week-by-week project schedule. Having a schedule for the project allowed me to concretely determine whether I was on-schedule or, if behind schedule, by how much. Starting the project by agreeing on a schedule also allowed me to be more intentional about how I carried out this project. I had read about the importance of human-centered design and about gathering user input in order build something useful. I wanted to make sure that this emphasis would be incorporated into the work.

User research on Reddit
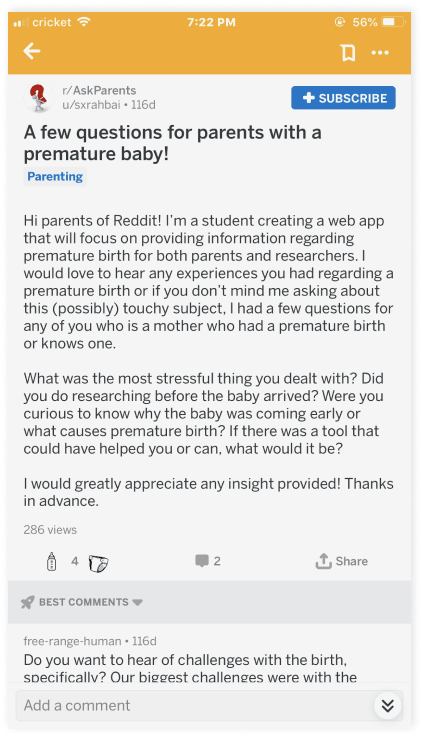
In addition to exploratory conversations with Dr. Sarkar about the researcher side of this project, I turned to Reddit support groups to reach out to mothers of premature newborns. I decided to use Reddit because of its engaged and active user base. I knew that if I posted on relevant sub-Reddits, I would likely get some response. For future work, I hope to be able to more systematically choose where I try to recruit people to speak to.
In each relevant Reddit support group, I posted an introductory blurb about my work and asked for open-ended feedback. For those who responded, I would invite them to continue our conversation over a 30-minute video chat session. Because of funding limitations, I wasn't able to compensate any of the people who generously donated their time to speak with me. In total, I was able to engage around a dozen mothers directly on Reddit and have follow-up conversations with two mothers.
Concept generation using Figma
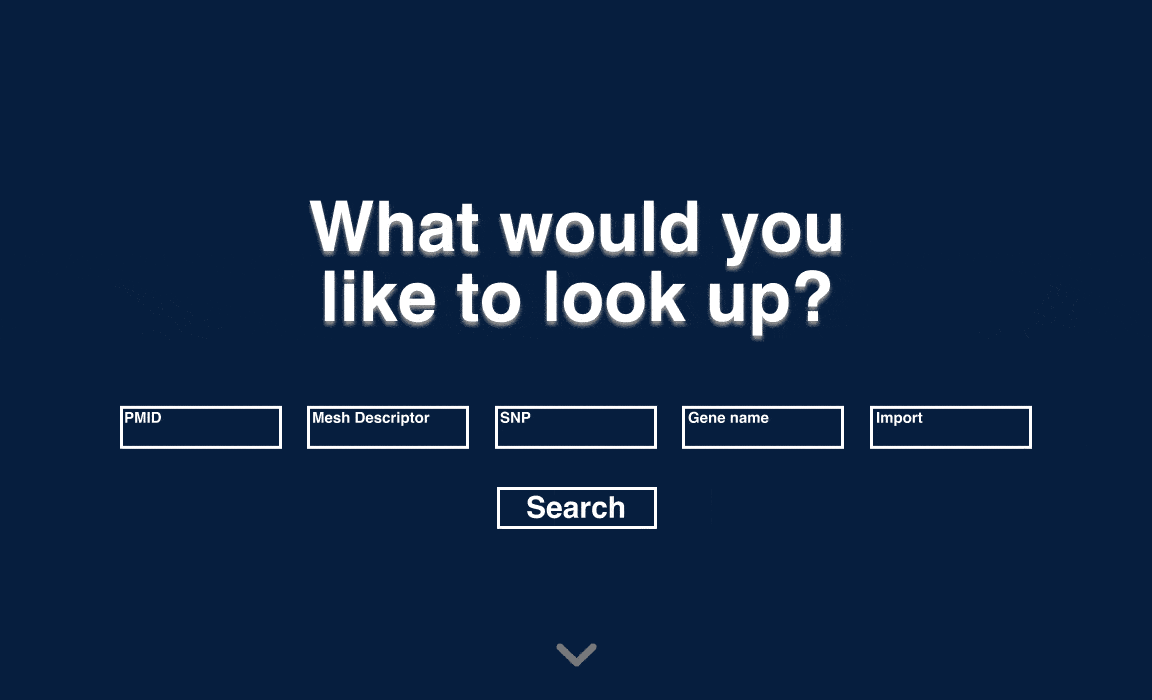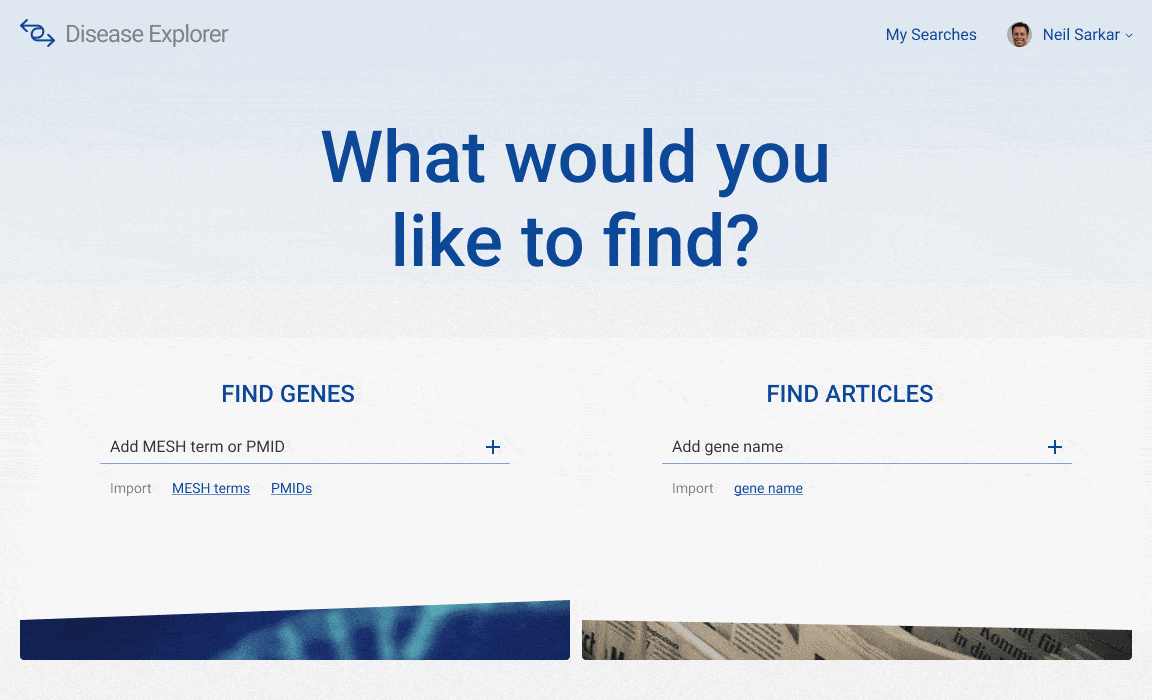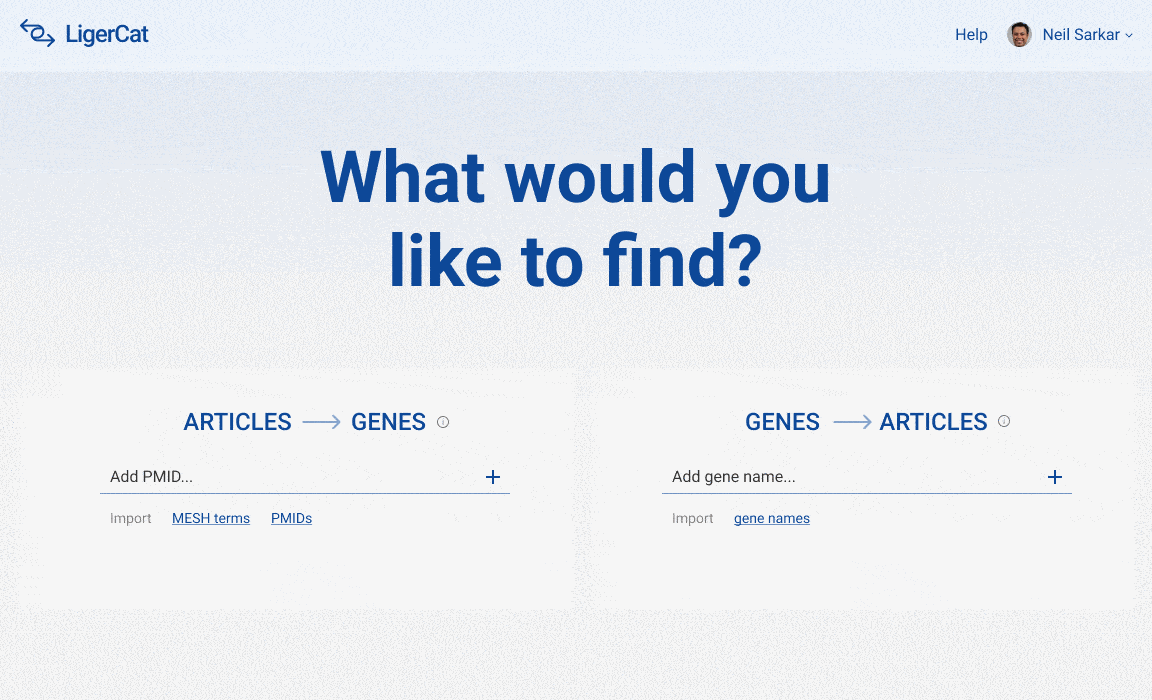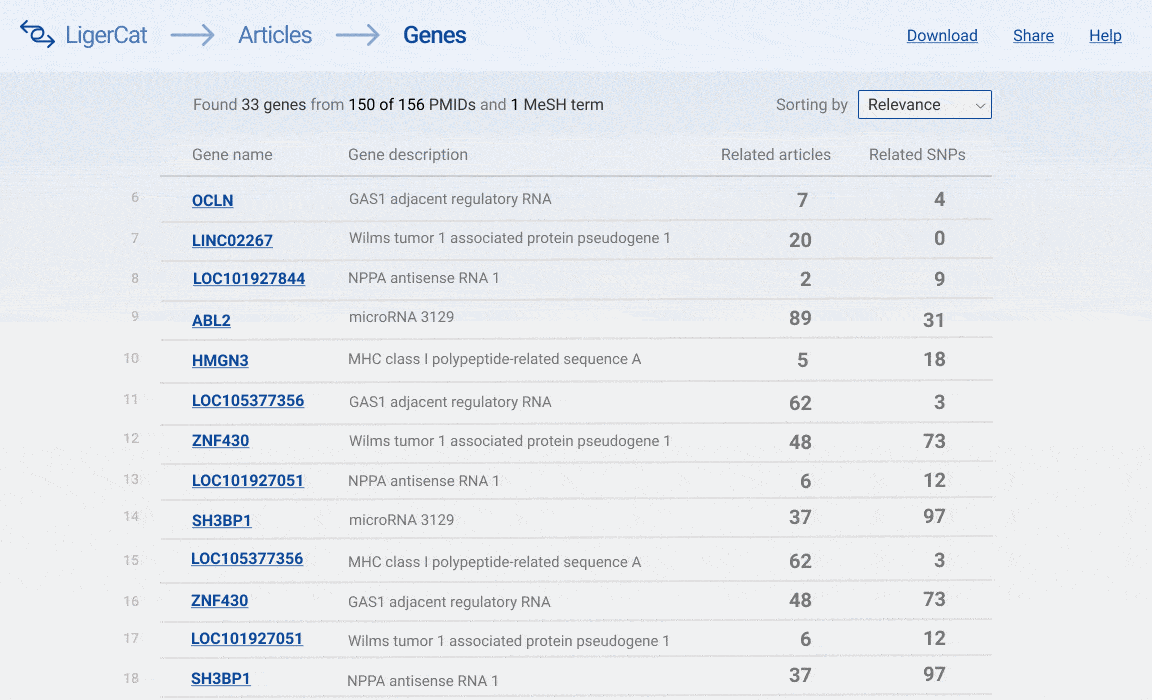
As I was doing informal user research, I started to develop concepts for LigerCat2's functionality and user interface. I experimented with different interfaces, from a browser extension that would modify the NCBI website to a standalone web interface. I decided to move forward with a standalone web interface because I wanted to aggregate data from multiple NCBI data sources and doing so in a cohesive single interface seemed most intuitive.
When deciding on the look and feel of the interface, I considered a dark versus a light theme. I ended up choosing to move forward with the lighter theme because I wanted the interface to emphasize accessibility and ease-of-use. I looked websites that might have similar goals -- primarily banking and government websites -- and settled on a similar lighter, more subdued palette.
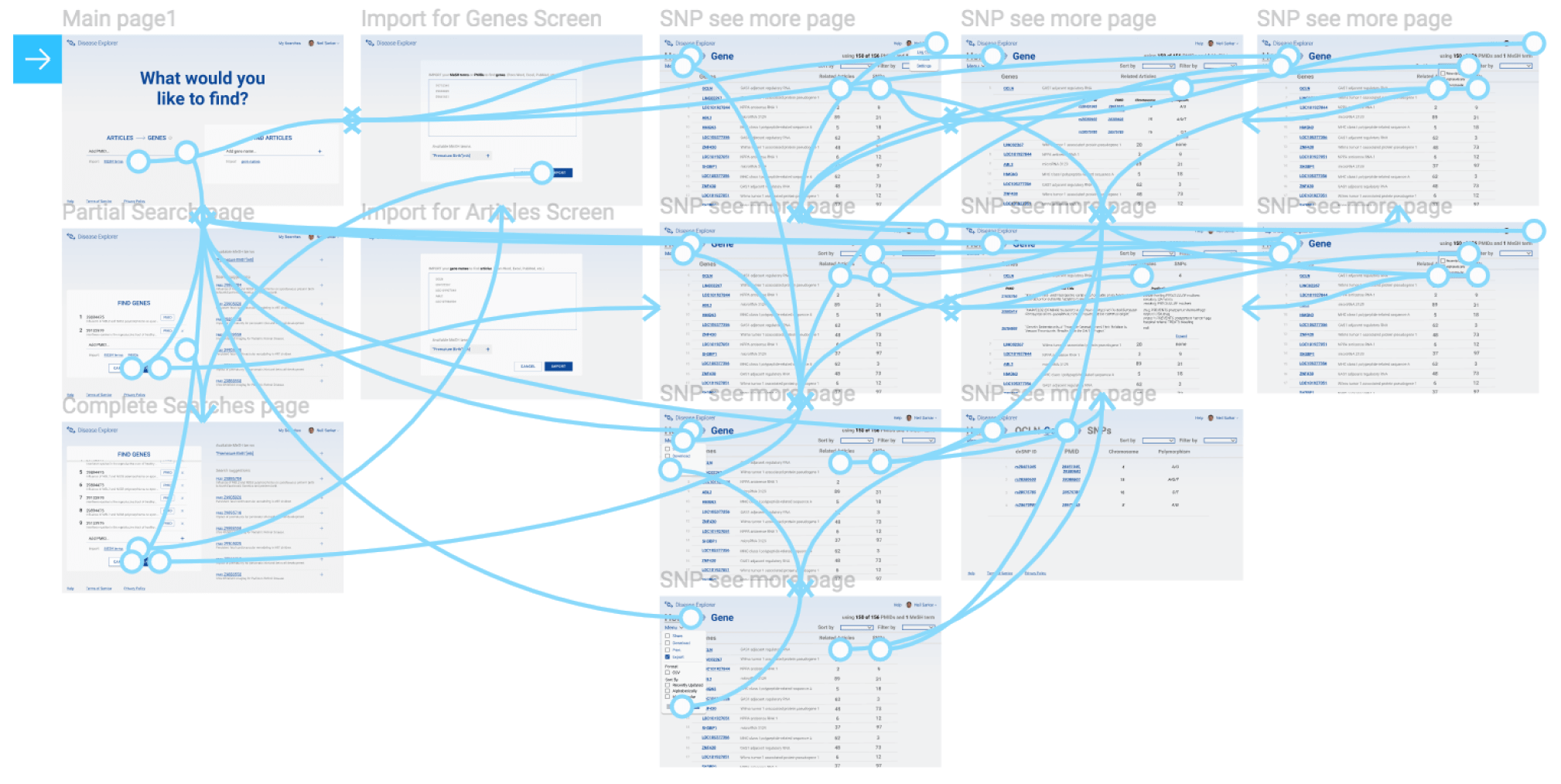
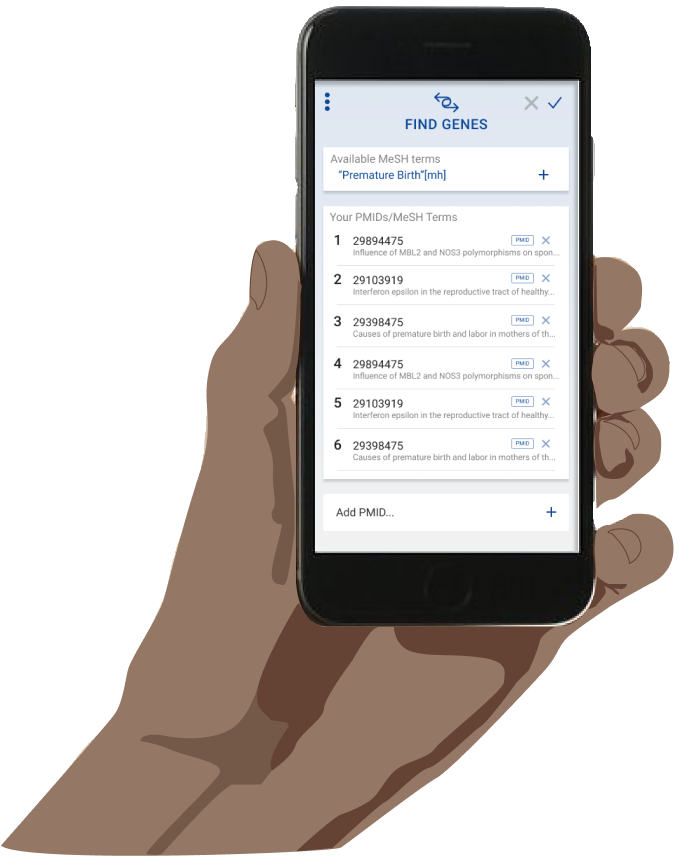
The final user interface is split into two halves: article to gene search on one side and gene to article search on the other. Using the prototyping functionality built into Figma, I linked together the various artboards of this user flow into a click through prototype. Because of time limitations, I was only able to review this click through prototype with Dr. Sarkar. Future work would be to gather more thorough user feedback on these mock-ups.
Iterative feedback
Throughout this entire process, I would meet weekly with Dr. Sarkar to review progress and ask any questions that came up during the week. Dr. Sarkar was especially helpful in guiding me through the more technical aspects of this project, including the code for extracting data from NCBI and developing the initial code prototype.
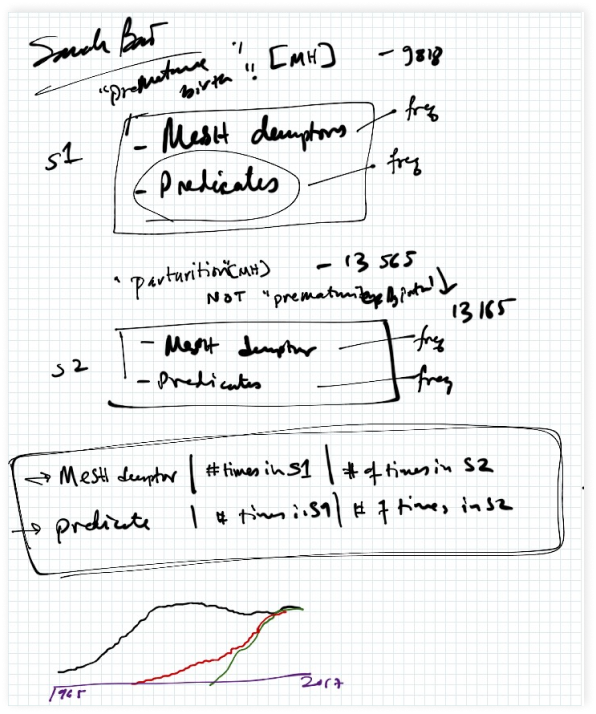
He also suggested several relevant resources for this project that I hope to include with future work. One such resource is the Semantic MEDLINE Database (SemMed), which is a database of semantically relevant sentence predications for all PubMed articles. I had hoped to use SemMed as the basis for developing the machine-generated accessible summaries of articles for mothers, but I was unable to fully flesh out this side of the project during the summer. Hopefully, I can continue to work with Dr. Sarkar to expand on this side of the project.
Data extraction using Julia
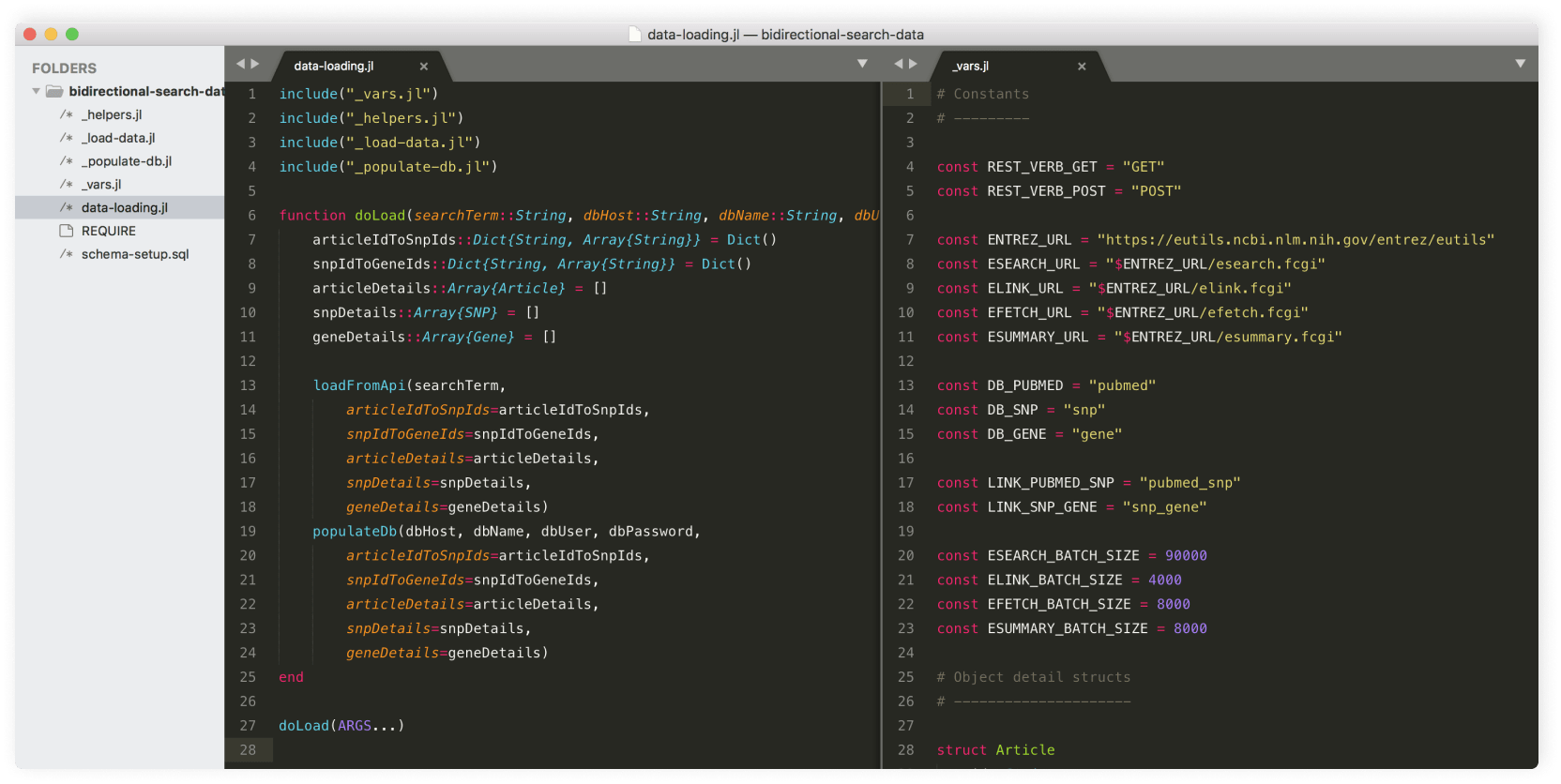
As I gradually arrived at a cohesive concept for this project, I turned my attention to gathering the data needed for this project. Building on the initial work I had done the previous summer, with Dr. Sarkar's help, I expanded the Julia scripts I had written to use the NCBI's Entrez API to build a MySQL database cross-referencing articles, SNPs, and genes related to prematurity.
First, I used ESearch to pull the article id, title, description, and journal name for all PubMed articles under the prematurity MeSH heading. Using these article ids, I looked up the SNP id, chromosome number, and observed bases for all corresponding SNPs in dbSNP using ELink. Using these SNP ids, I used ESummary to obtain the gene id, name, chromosome location, and description for all relevant genes. Once all of the article, SNP, and gene information was gathered, I saved all of this information in a MySQL database for use in the code prototype.
In the future, I hope to extend this script to run periodically so that newly added articles, SNPs, and genes will be included in the LigerCat2 database.
Prototype development using Django
After conducting initial user research, developing concepts based on user feedback, and extracting the relevant data, the last part of this project was to create a working code prototype for LigerCat2. This was the most challenging part of the project for me because, while I had some Julia experience from taking Dr. Sarkar's class last summer and some design experience from earlier work, I hadn't built a web application before.
I used Django to build this initial code prototype because it seemed to be the easiest to set up and had really good documentation. I leaned heavily on the tutorials I found on the Django Girls website to learn the basics of Django, including how to connect Django to my database, how to build webpages in Django, and how to link these webpages together.
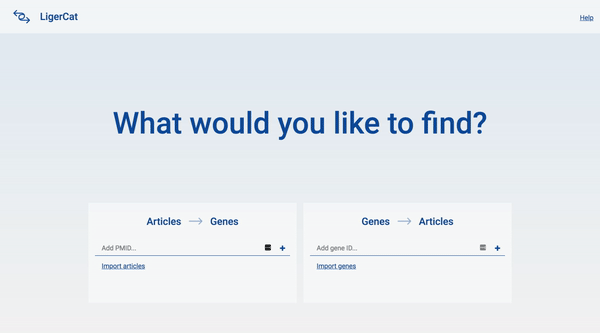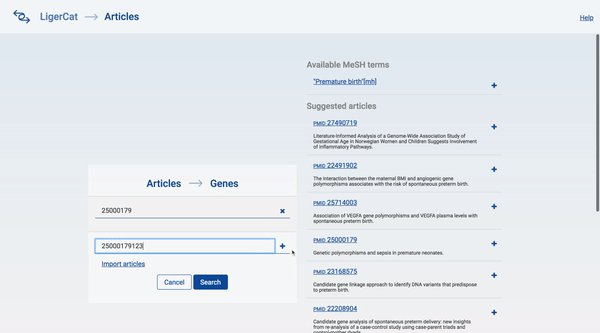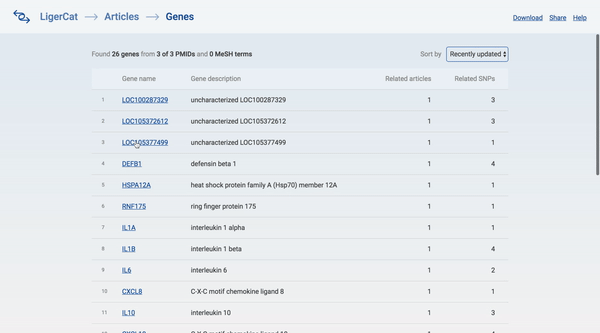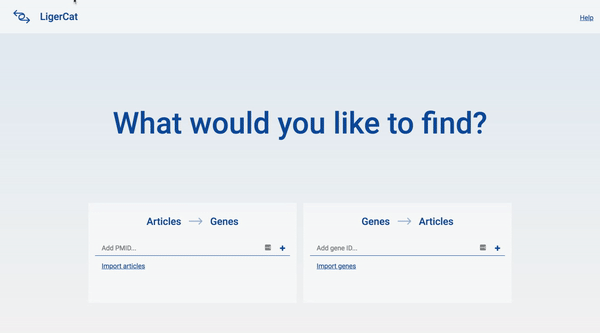
In the end, I was able to flesh out the basic articles to genes workflow for the desktop interface. A user can type in a series of article ids and get a list of real, relevant gene search results. The user can then click into any gene search result to see detailed information about that gene and access direct links to relevant NCBI pages.
Going forward I hope to make the user interface more intuitive by allowing users to search by article titles, flesh out the sharing and downloading functionalities, flesh out the reverse genes to articles workflow, and create a mobile interface for LigerCat2. Also, while I was able to conduct some initial research on how patients access prematurity information and to formulate initial concepts on how to improve access, I wasn't able to integrate or implement any of these concepts. The patient side of this project is important to me and something that I want to expand going forward.
LigerCat2
Working with Dr. Sarkar on LigerCat2 was a treat. I was able to touch on all aspects of creating a product, from ideation to user research to concept generation to prototyping. I enjoyed using Figma to render ideas in higher fidelity than just sketching on a piece of paper. Figma's prototyping capabilities also allowed me to create clickable mock-ups that were much more engaging than simply viewing static screens. I expanded my Julia programming skills and was able to take a first pass at building a web application.
Most importantly, I was able to see the how a medical condition has multiple sides and to find the common desire that unites these perspectives: a shared need for accessible information. In the case of prematurity, both researchers and patients need high-quality information because they both want better treatment and outcomes. I hope to continue to work on using thoughtful research and design to build solutions for bridging these gaps.